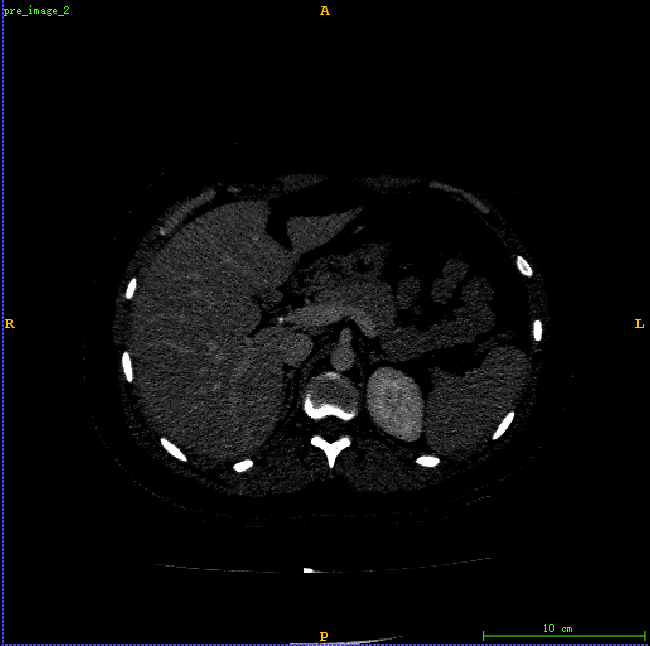
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
| import torch
import torch.nn as nn
import SimpleITK as sitk
import numpy as np
def change_indenty(ct):
ct[ct < 40] = 40
ct[ct > 400] = 400
return ct
class Dialte(nn.Module):
def __init__(self, num):
super(Dialte, self).__init__()
self.num= num
self.act = nn.ReLU(inplace=False)
self.norm = nn.BatchNorm3d
self.conv1 = nn.Sequential(
nn.Conv3d(1,1, kernel_size=3, stride=1, padding=1),
self.act,
self.norm(1)
)
def forward(self, x):
for i in range(self.num):
x = self.conv1(x)
return x
if __name__ == "__main__":
path = r"D:\myProject\HDC_vessel_seg\datasets\nii\image_2.nii"
image = sitk.ReadImage(path)
img_num = sitk.GetArrayFromImage(image)
img_num = change_indenty(img_num)
img_num = np.expand_dims(np.expand_dims(img_num, axis=0), axis=0).astype(np.float32)
img_num = torch.from_numpy(img_num)
print(img_num.shape)
model = Dialte(num=5)
x = model(img_num)
x = x[0,0,...]
x = x.cpu().data.numpy()
print("x",x.shape)
predict_seg = sitk.GetImageFromArray(x)
predict_seg.SetSpacing(image.GetSpacing())
predict_seg.SetOrigin(image.GetOrigin())
predict_seg.SetDirection(image.GetDirection())
sitk.WriteImage(predict_seg, path.replace("image", "pre_image"))
|